It’s good practice to normalize the different numeric variables
Code
d=iris[,-5] # numerical iris data (without speciee) ds=scale(d) # scaled iris data (column-wise)br=seq(-5,7,by=0.5) # set common break points for the histograms belowhist(d[,"Sepal.Width"], breaks = br) # illustrate scaling for specific columnhist(ds[,"Sepal.Width"], breaks = br, add=TRUE, col="red") # add histogram to current plotlegend("topright", c("orig","scaled"), fill=c("white", "red"))
Heatmaps
Heatmaps are color-coded representations of numerical matrices.
Typically the rows and columns are re-ordered according to some distance measure (default: Euclidean) and hierarchical clustering method (default: complete)
There are many tools to draw heatmaps in R. Here we use the pheatmap package to provide this powerful functionality
Code
#install.packages("pheatmap") # That's how we install new packages - more laterlibrary(pheatmap) # make packaged functions availablepaste('loaded pheatmap version:', packageVersion('pheatmap'))
[1] "loaded pheatmap version: 1.0.12"
Code
ds =scale(iris[,-5]) # scaled data for heatmapann =data.frame(Species = iris[,5]) # meta data for annotations# explicitly set rownames to retain association between data and metadatarownames(ds)=rownames(iris)rownames(ann)=rownames(iris)pheatmap(ds, annotation_row = ann,show_rownames =FALSE, )
There is many more parameters for more control - if you have lots of time read “?pheatmap”
Sending plots to files
In Rstudio, we can export figures from the “Plots” tab. On the console we can define a pdf file as a new device for all subsequent figures. This is usually done only after the image is sufficiently optimized
Code
pdf("output/heatmap.pdf") # similar for jpeg, png, ...pheatmap(ds, annotation_row = ann, show_rownames =FALSE)dev.off() # close device = pdf file
pdf
3
Goal 2: Show me all the data (in lower dimensions)
Code
M=as.matrix(iris[,-5]) # numerical data, some operations below require matrices not data frames (%*%)s=iris[,5] # species attributes (factor)
PCA Goals
simplify description of data matrix \(M\): data reduction & extract most important information
maximal variance: look for direction in which data shows maximal variation
minimal error: allow accurate reconstruction of original data
from amoeba @ https://stats.stackexchange.com/questions/2691/making-sense-of-principal-component-analysis-eigenvectors-eigenvalues
For the iris data, there are many correlations
Code
pheatmap(cor(M)) # original correlation matrix
PCA with R
Code
pca =prcomp(M, scale=TRUE)
Task
What kind of object is pca?
Correlation Structure
Code
S=pca$x # score matrix = rotated and scaled data matrixpairs(S, col=s)
Code
# correlation matrix of scores is diagonal (by design)# --> principal components are uncorrelatedpheatmap(cor(S))
Ordered Components
First principle components contribute most to the overall variance
Code
plot(pca)
The success of this procedure depends on the data, but if there is a strong ordering, then we might represent the data in only the first two dimensions:
Code
plot(S[,1:2],pch=21, bg=s) # score-plot
Review
use heatmaps to visualize large matrices
data transformation: scale()
installing packages: pheatmap
exporting figures as publication-ready files: pdf()
dimensional reduction (PCA): prcomp()
Source Code
---title: "06: Data Visualization"author: "Thomas Manke"categories: - pheatmap - PCA---```{r, child="_setup.qmd"}```## Goal 1: Show me all the data### Scaling It's good practice to normalize the different numeric variables```{r scaling}d=iris[,-5] # numerical iris data (without speciee) ds=scale(d) # scaled iris data (column-wise)br=seq(-5,7,by=0.5) # set common break points for the histograms belowhist(d[,"Sepal.Width"], breaks = br) # illustrate scaling for specific columnhist(ds[,"Sepal.Width"], breaks = br, add=TRUE, col="red") # add histogram to current plotlegend("topright", c("orig","scaled"), fill=c("white", "red"))```### HeatmapsHeatmaps are color-coded representations of numerical matrices. Typically the rows and columns are re-ordered according to some distance measure (default: Euclidean) and hierarchical clustering method (default: complete) There are many tools to draw heatmaps in R.Here we use the `pheatmap` package to provide this powerful functionality```{r pheatmap}#install.packages("pheatmap") # That's how we install new packages - more laterlibrary(pheatmap) # make packaged functions availablepaste('loaded pheatmap version:', packageVersion('pheatmap'))ds = scale(iris[,-5]) # scaled data for heatmapann = data.frame(Species = iris[,5]) # meta data for annotations# explicitly set rownames to retain association between data and metadatarownames(ds)=rownames(iris)rownames(ann)=rownames(iris)pheatmap(ds, annotation_row = ann, show_rownames = FALSE, )```There is many more parameters for more control - if you have lots of time read "?pheatmap" ***### Sending plots to filesIn Rstudio, we can export figures from the "Plots" tab. On the console we can define a pdf file as a new device for all subsequent figures. This is usually done only *after* the image is sufficiently optimized```{r pdf}pdf("output/heatmap.pdf") # similar for jpeg, png, ...pheatmap(ds, annotation_row = ann, show_rownames = FALSE)dev.off() # close device = pdf file```***## Goal 2: Show me all the data (in lower dimensions)```{r df2mat}M=as.matrix(iris[,-5]) # numerical data, some operations below require matrices not data frames (%*%)s=iris[,5] # species attributes (factor)```### PCA Goals* simplify description of data matrix $M$: data reduction & extract most important information* maximal variance: look for direction in which data shows maximal variation* minimal error: allow accurate reconstruction of original data 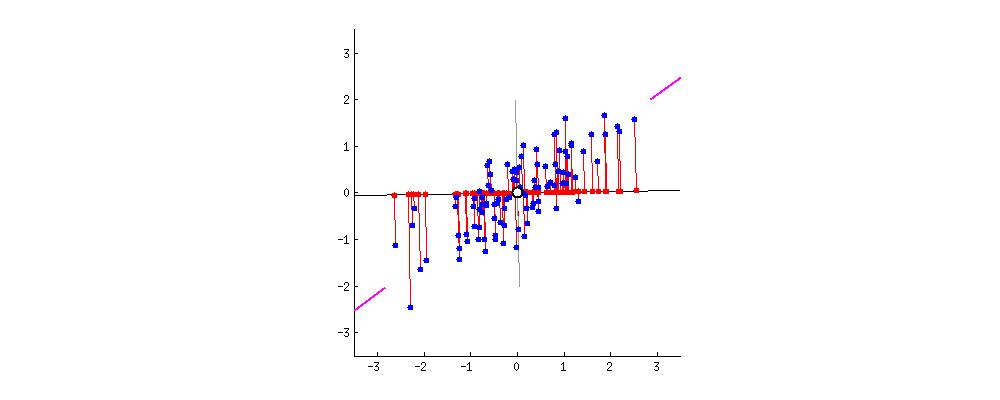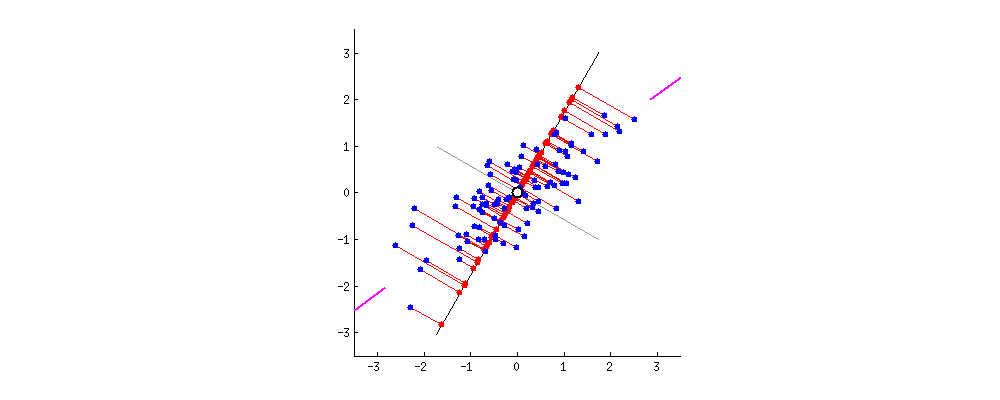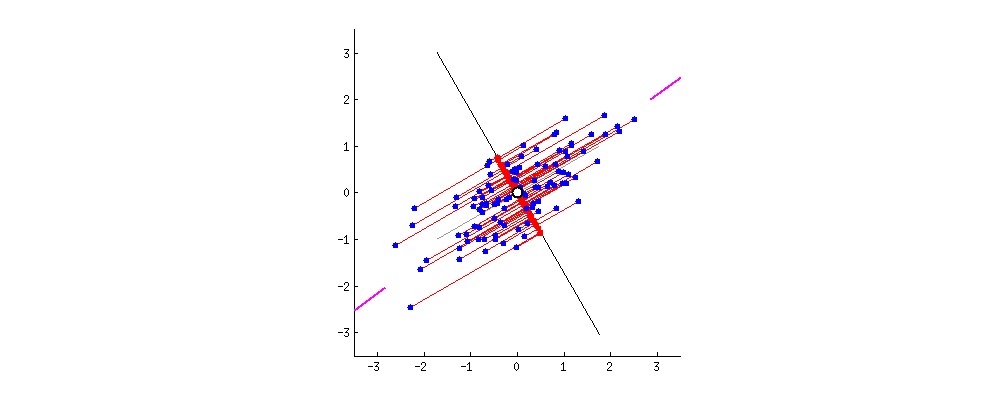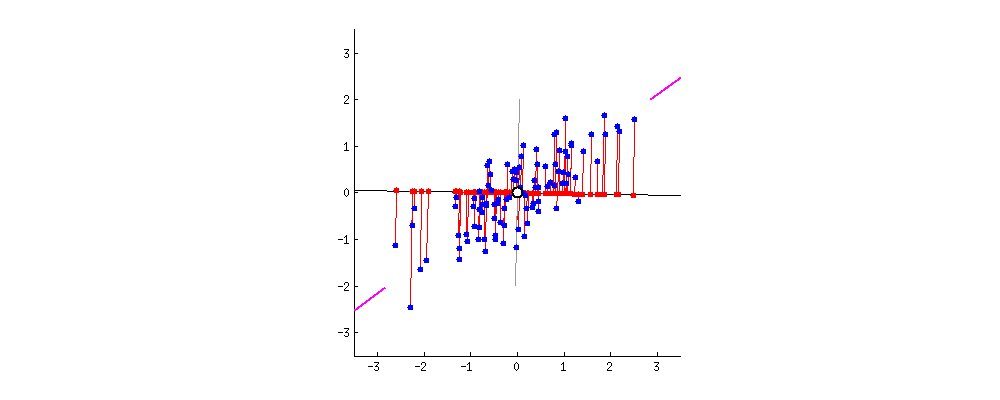from amoeba @ https://stats.stackexchange.com/questions/2691/making-sense-of-principal-component-analysis-eigenvectors-eigenvaluesFor the iris data, there are many correlations```{r}pheatmap(cor(M)) # original correlation matrix```### PCA with R```{r run_pca}pca = prcomp(M, scale=TRUE)```#### Task What kind of object is pca?```{r pca_obj, echo=FALSE, eval=FALSE}class(pca)typeof(pca)str(pca)methods(class="prcomp")```### Correlation Structure```{r pca_cov}S=pca$x # score matrix = rotated and scaled data matrixpairs(S, col=s)# correlation matrix of scores is diagonal (by design)# --> principal components are uncorrelatedpheatmap(cor(S)) ```### Ordered ComponentsFirst principle components contribute most to the overall variance```{r}plot(pca)```The success of this procedure depends on the data, but **if** there is a strong ordering, then we might represent the data in only the **first two dimensions**:```{r plot_PC1_PC2}plot(S[,1:2],pch=21, bg=s) # score-plot```***## Review* use heatmaps to visualize large matrices* data transformation: scale()* installing packages: pheatmap* exporting figures as publication-ready files: pdf()* dimensional reduction (PCA): prcomp()